A growing body of evidence supports the hypothesis that amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) share underlying mechanisms and are placed on the opposite ends of a disease spectrum that leads to these pathologies.
This connection is supported by the initial discovery that two different RNA binding proteins, TDP-43 and FUS/TLS, form neuronal inclusions in neurons of ALS and FTD patients.
In addition, autosomal dominant forms of FTD, ALS, or a combined phenotype can occur in the same family.
Further genetic studies have found an association between an expanded GGGGCC hexanucleotide repeat in the C9ORF72 gene and both familiar and sporadic cases of FTD and ALS (namely, c9FTD/ALS)
Several reports suggest that the increased nucleotide repeats in C9ORF72 causes the formation of RNA inclusions or of foci in the nucleus of affected cells.
Apparently, the repeats expansion in C9ORF72 can be pathogenic by sequestering RNA binding proteins and/or perturbing both splicing and regulation of key factors implicated in neuronal metabolisms.
It is interesting to note that TDP-43 and FUS/TLS play fundamental roles in RNA regulation and splicing, and their mutation causes alteration of these processes.
Altogether these findings suggest that defects in RNA metabolism can be a common pathway linking FTD to ALS, and are responsible for disease pathogenesis in a significant number of cases.
Studies aimed at testing this hypothesis and determine the pathways regulated by these proteins will be helpful in improved understanding of the molecular mechanisms underlying the pathogenesis of ALS and FTD, ultimately opening up new routes of therapy for these devastating disorders.
Keywords
Splicing, TDP-43, proteinopathy, neurodegeneration, inclusion AD, ALS, FTD, FTLD, FUS, TLS, C9ORF72, PD.
Ongoing Projects
The actual studies of my research group are aimed at molecular dissection of the mechanisms controlling pre-mRNA processing in physiological and pathological conditions, with a special interest in neurodegenerative diseases.
These aims are pursued by using in vitro and ex vivo approaches, complemented by transcriptomic and computational techniques, to carry out molecular characterization of pathophysiologic events and to grasp a global view of the underlying regulatory networks.
The current research of the laboratory centres around:
- TDP-43 functions and pathogenic mechanisms involved in TDP-43 proteinopathies.
- Splicing alterations and genetic diseases.
- SigmaR1 and SigmaR2 ligands: characterization of their effects and possible use for innovative therapies.
- Role of cholinergic and noradrenergic transmitter systems in the development of cognitive deficits, as well as in the occurrence of histopathological alterations (e.g. amyloid, tau, TDP-43 or alpha-synuclein overexpression) in relevant brain areas.
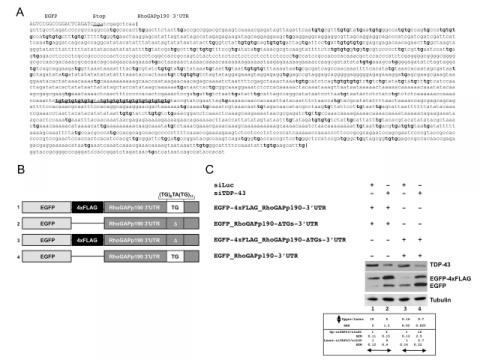
Figure - TDP-43 is implicated in the regulation of RhoGAPp190 mRNA steady-state level
A) Sequence of the 3’UTR of RhoGAPp190 cloned in the pEGFP-C2 vector. All TG dinucleotide repeats within the 3'UTR are boldface. The TG-repeat rich sequence located 1470nts downstream of the stop codon is underlined. B) Schematic representation of the EGFP- RhoGAPp190-3’UTR constructs. All relevant elements within the construct are shown. C) Effect of TDP-43 depletion on EGFP-RhoGAPp190-3’UTR constructs. Equal amounts of the indicated EGFP-RhoGAPp190 constructs were co-transfected in Hek293 cells. The effects of TDP-43 silencing (siTDP-43) on EGFP expression were monitored by anti-GFP immunoblotting. Anti-luciferase siRNA (siLUC) was used as the control. Western blot anti-tubulin (Tubulin) served as the protein loading control. One representative blot is shown of three independent experiments. Quantification of protein levels are shown as relative ratio of upper vs lower band in each condition or of siTDP vs siLUC treatment. Values are expressed in mean (of three independent experiments) ± SEM. P<0.05. (Langellotti S, Romano G, Feiguin F, Baralle FE, Romano M (2018) RhoGAPp190: A potential player in tbph-mediated neurodegeneration in Drosophila. PLoS ONE 13(4): e0195845. https://doi.org/10.1371/journal.pone.0195845)




